Info
10x_
10x Genomics (2022)
72.38 MiB
20-08-2024
3460 × 13565
Gene expression library of Human Prostate Cancer (Visium FFPE) with an IF image using the Human Whole Transcriptome Probe Set
10x_
10x Genomics (2022)
72.38 MiB
20-08-2024
3460 × 13565
CREATED
20-08-2024
DIMENSIONS
3460 × 13565
10x Genomics obtained FFPE human prostate tissue from Indivumed Human Tissue Specimens. Original diagnosis with adenocarcinoma. The tissue was sectioned as described in Visium Spatial Gene Expression for FFPE Tissue Preparation Guide Demonstrated Protocol (CG000408). Tissue sections of 10 µm were placed on Visium Gene Expression slides, then stained following Deparaffinization, Decrosslinking, Immunofluorescence Staining & Imaging Demonstrated Protocol (CG000410).
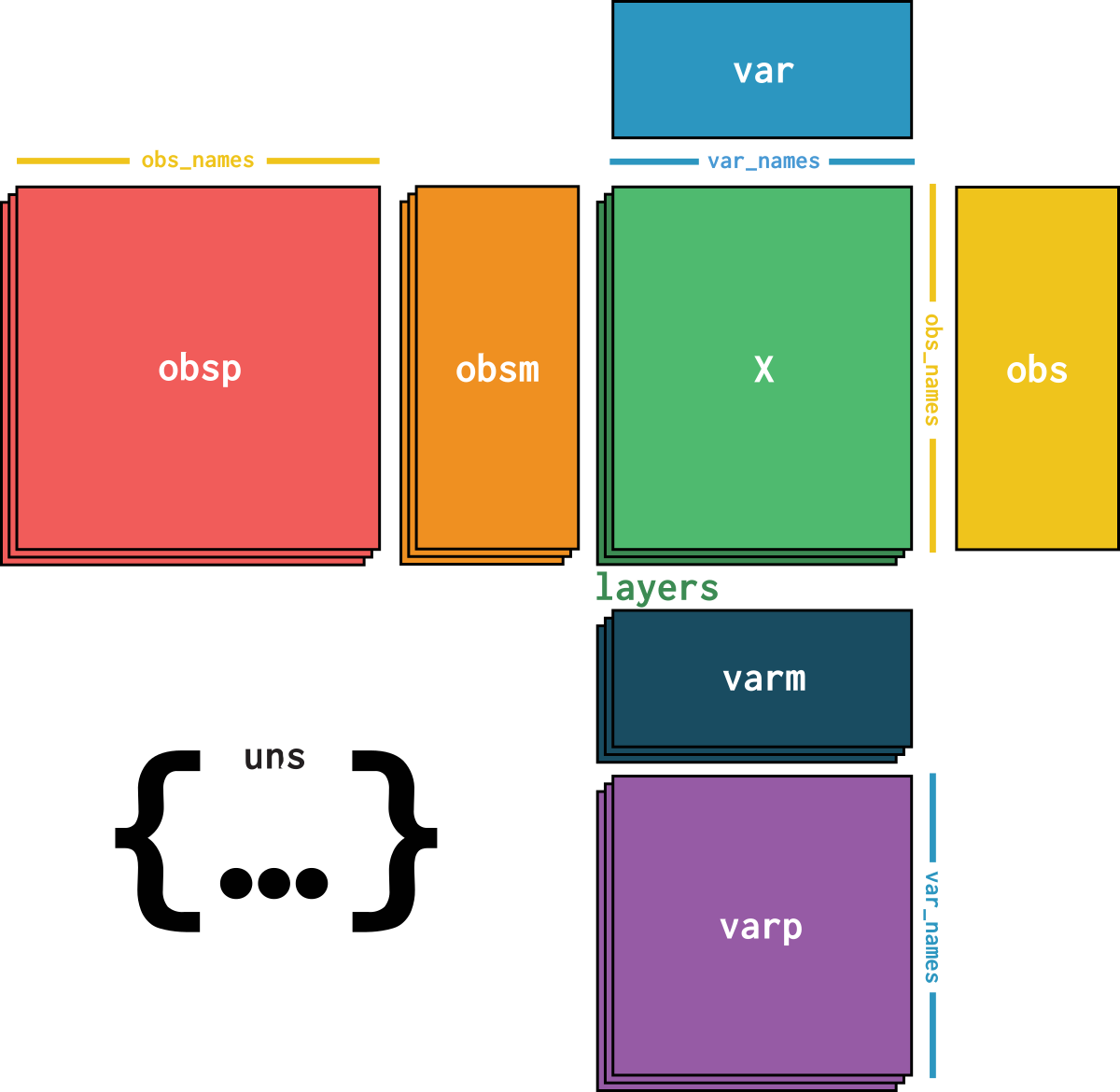
dataset is an AnnData object with n_obs × n_vars = 3460 × 13565 with slots:
feature_id, feature_namecountsdataset_description, dataset_id, dataset_name, dataset_organism, dataset_reference, dataset_summary, dataset_url| Name | Description | Type | Data type | Size |
|---|---|---|---|---|
| var | ||||
feature_
|
Unique identifier for the feature, usually a ENSEMBL gene id. |
vector
|
object
|
13565 |
feature_
|
A human-readable name for the feature, usually a gene symbol. |
vector
|
object
|
13565 |
| layers | ||||
counts
|
Raw counts |
sparsematrix
|
float32
|
3460 × 13565 |
| uns | ||||
dataset_
|
Long description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
|
atomic
|
str
|
1 |
dataset_
|
A human-readable name for the dataset. |
atomic
|
str
|
1 |
dataset_
|
The organism of the sample in the dataset. |
atomic
|
str
|
1 |
dataset_
|
Bibtex reference of the paper in which the dataset was published. |
atomic
|
str
|
1 |
dataset_
|
Short description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
Link to the original source of the dataset. |
atomic
|
str
|
1 |
dataset.layers['counts']In R: dataset$layers[["counts"]]
Type: sparsematrix, data type: float32, shape: 3460 × 13565
Raw counts
dataset.uns['dataset_description']In R: dataset$uns[["dataset_description"]]
Type: atomic, data type: str, shape: 1
Long description of the dataset.
dataset.uns['dataset_id']In R: dataset$uns[["dataset_id"]]
Type: atomic, data type: str, shape: 1
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
dataset.uns['dataset_name']In R: dataset$uns[["dataset_name"]]
Type: atomic, data type: str, shape: 1
A human-readable name for the dataset.
dataset.uns['dataset_organism']In R: dataset$uns[["dataset_organism"]]
Type: atomic, data type: str, shape: 1
The organism of the sample in the dataset.
dataset.uns['dataset_reference']In R: dataset$uns[["dataset_reference"]]
Type: atomic, data type: str, shape: 1
Bibtex reference of the paper in which the dataset was published.
dataset.uns['dataset_summary']In R: dataset$uns[["dataset_summary"]]
Type: atomic, data type: str, shape: 1
Short description of the dataset.
dataset.uns['dataset_url']In R: dataset$uns[["dataset_url"]]
Type: atomic, data type: str, shape: 1
Link to the original source of the dataset.
dataset.var['feature_id']In R: dataset$var[["feature_id"]]
Type: vector, data type: object, shape: 13565
Unique identifier for the feature, usually a ENSEMBL gene id.
dataset.var['feature_name']In R: dataset$var[["feature_name"]]
Type: vector, data type: object, shape: 13565
A human-readable name for the feature, usually a gene symbol.