Info
cellxgene_
Steuernagel et al. (2022)
12.58 GiB
02-02-2024
384925 × 41642
Quick links
Used in
No related benchmarks found.
A unified single cell gene expression atlas of the murine hypothalamus
CREATED
02-02-2024
DIMENSIONS
384925 × 41642
The hypothalamus plays a key role in coordinating fundamental body functions. Despite recent progress in single-cell technologies, a unified catalogue and molecular characterization of the heterogeneous cell types and, specifically, neuronal subtypes in this brain region are still lacking. Here we present an integrated reference atlas “HypoMap” of the murine hypothalamus consisting of 384,925 cells, with the ability to incorporate new additional experiments. We validate HypoMap by comparing data collected from SmartSeq2 and bulk RNA sequencing of selected neuronal cell types with different degrees of cellular heterogeneity.
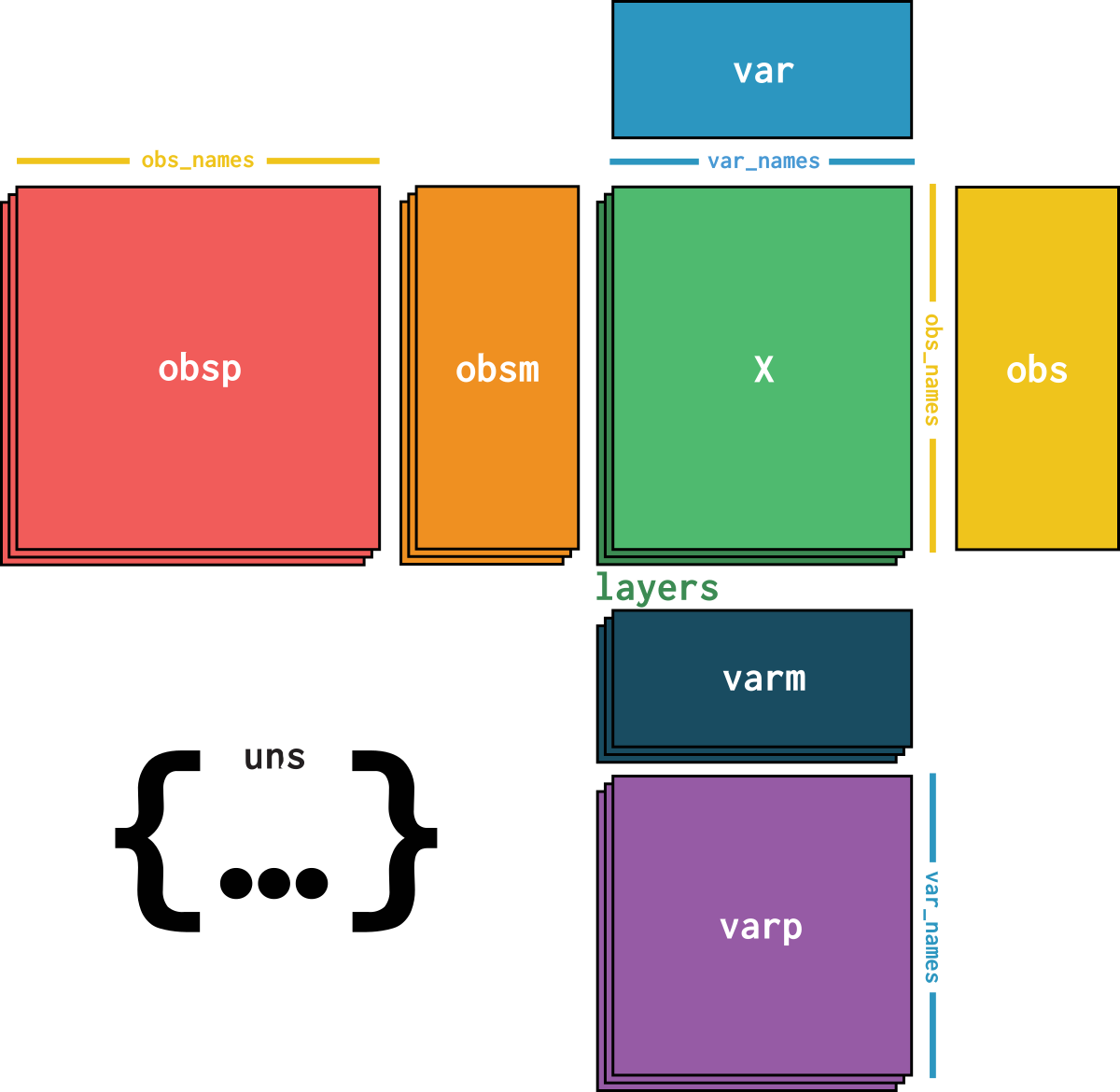
dataset is an AnnData object with n_obs × n_vars = 384925 × 41642 with slots:
soma_joinid, dataset_id, assay, assay_ontology_term_id, cell_type, cell_type_ontology_term_id, development_stage, development_stage_ontology_term_id, disease, disease_ontology_term_id, donor_id, is_primary_data, self_reported_ethnicity, self_reported_ethnicity_ontology_term_id, sex, sex_ontology_term_id, suspension_type, tissue, tissue_ontology_term_id, tissue_general, tissue_general_ontology_term_id, batch, size_factorssoma_joinid, feature_id, feature_name, hvg, hvg_scoreknn_connectivities, knn_distancesX_pcapca_loadingscounts, normalizeddataset_description, dataset_id, dataset_name, dataset_organism, dataset_reference, dataset_summary, dataset_url, knn, normalization_id, pca_variance| Name | Description | Type | Data type | Size |
|---|---|---|---|---|
| obs | ||||
assay
|
Type of assay used to generate the cell data, indicating the methodology or technique employed. |
vector
|
category
|
384925 |
assay_
|
Experimental Factor Ontology (EFO:) term identifier for the assay, providing a standardized reference to the assay type.
|
vector
|
category
|
384925 |
batch
|
A batch identifier. This label is very context-dependent and may be a combination of the tissue, assay, donor, etc. |
vector
|
category
|
384925 |
cell_
|
Classification of the cell type based on its characteristics and function within the tissue or organism. |
vector
|
category
|
384925 |
cell_
|
Cell Ontology (CL:) term identifier for the cell type, offering a standardized reference to the specific cell classification.
|
vector
|
category
|
384925 |
dataset_
|
Identifier for the dataset from which the cell data is derived, useful for tracking and referencing purposes. |
vector
|
category
|
384925 |
development_
|
Stage of development of the organism or tissue from which the cell is derived, indicating its maturity or developmental phase. |
vector
|
category
|
384925 |
development_
|
Ontology term identifier for the developmental stage, providing a standardized reference to the organism’s developmental phase. If the organism is human (organism_ontology_term_id == 'NCBITaxon:9606'), then the Human Developmental Stages (HsapDv:) ontology is used. If the organism is mouse (organism_ontology_term_id == 'NCBITaxon:10090'), then the Mouse Developmental Stages (MmusDv:) ontology is used. Otherwise, the Uberon (UBERON:) ontology is used.
|
vector
|
category
|
384925 |
disease
|
Information on any disease or pathological condition associated with the cell or donor. |
vector
|
category
|
384925 |
disease_
|
Ontology term identifier for the disease, enabling standardized disease classification and referencing. Must be a term from the Mondo Disease Ontology (MONDO:) ontology term, or PATO:0000461 from the Phenotype And Trait Ontology (PATO:).
|
vector
|
category
|
384925 |
donor_
|
Identifier for the donor from whom the cell sample is obtained. |
vector
|
category
|
384925 |
is_
|
Indicates whether the data is primary (directly obtained from experiments) or has been computationally derived from other primary data. |
vector
|
bool
|
384925 |
self_
|
Ethnicity of the donor as self-reported, relevant for studies considering genetic diversity and population-specific traits. |
vector
|
category
|
384925 |
self_
|
Ontology term identifier for the self-reported ethnicity, providing a standardized reference for ethnic classifications. If the organism is human (organism_ontology_term_id == 'NCBITaxon:9606'), then the Human Ancestry Ontology (HANCESTRO:) is used.
|
vector
|
category
|
384925 |
sex
|
Biological sex of the donor or source organism, crucial for studies involving sex-specific traits or conditions. |
vector
|
category
|
384925 |
sex_
|
Ontology term identifier for the biological sex, ensuring standardized classification of sex. Only PATO:0000383, PATO:0000384 and PATO:0001340 are allowed.
|
vector
|
category
|
384925 |
size_
|
The size factors created by the normalisation method, if any. |
vector
|
float32
|
384925 |
soma_
|
If the dataset was retrieved from CELLxGENE census, this is a unique identifier for the cell. |
vector
|
int64
|
384925 |
suspension_
|
Type of suspension or medium in which the cells were stored or processed, important for understanding cell handling and conditions. |
vector
|
category
|
384925 |
tissue
|
Specific tissue from which the cells were derived, key for context and specificity in cell studies. |
vector
|
category
|
384925 |
tissue_
|
General category or classification of the tissue, useful for broader grouping and comparison of cell data. |
vector
|
category
|
384925 |
tissue_
|
Ontology term identifier for the general tissue category, aiding in standardizing and grouping tissue types. For organoid or tissue samples, the Uber-anatomy ontology (UBERON:) is used. The term ids must be a child term of UBERON:0001062 (anatomical entity). For cell cultures, the Cell Ontology (CL:) is used. The term ids cannot be CL:0000255, CL:0000257 or CL:0000548.
|
vector
|
category
|
384925 |
tissue_
|
Ontology term identifier for the tissue, providing a standardized reference for the tissue type. For organoid or tissue samples, the Uber-anatomy ontology (UBERON:) is used. The term ids must be a child term of UBERON:0001062 (anatomical entity). For cell cultures, the Cell Ontology (CL:) is used. The term ids cannot be CL:0000255, CL:0000257 or CL:0000548.
|
vector
|
category
|
384925 |
| var | ||||
feature_
|
Unique identifier for the feature, usually a ENSEMBL gene id. |
vector
|
object
|
41642 |
feature_
|
A human-readable name for the feature, usually a gene symbol. |
vector
|
object
|
41642 |
hvg
|
Whether or not the feature is considered to be a ‘highly variable gene’ |
vector
|
bool
|
41642 |
hvg_
|
A ranking of the features by hvg. |
vector
|
float64
|
41642 |
soma_
|
If the dataset was retrieved from CELLxGENE census, this is a unique identifier for the feature. |
vector
|
int64
|
41642 |
| obsp | ||||
knn_
|
K nearest neighbors connectivities matrix. |
sparsematrix
|
float32
|
384925 × 384925 |
knn_
|
K nearest neighbors distance matrix. |
sparsematrix
|
float64
|
384925 × 384925 |
| obsm | ||||
X_
|
The resulting PCA embedding. |
densematrix
|
float32
|
384925 × 50 |
| varm | ||||
pca_
|
The PCA loadings matrix. |
densematrix
|
float32
|
41642 × 50 |
| layers | ||||
counts
|
Raw counts |
sparsematrix
|
float32
|
384925 × 41642 |
normalized
|
Normalised expression values |
sparsematrix
|
float32
|
384925 × 41642 |
| uns | ||||
dataset_
|
Long description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
|
atomic
|
str
|
1 |
dataset_
|
A human-readable name for the dataset. |
atomic
|
str
|
1 |
dataset_
|
The organism of the sample in the dataset. |
atomic
|
str
|
1 |
dataset_
|
Bibtex reference of the paper in which the dataset was published. |
atomic
|
str
|
1 |
dataset_
|
Short description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
Link to the original source of the dataset. |
atomic
|
str
|
1 |
knn
|
Supplementary K nearest neighbors data. |
dict
|
3 | |
normalization_
|
Which normalization was used |
atomic
|
str
|
1 |
pca_
|
The PCA variance objects. |
dict
|
2 | |
dataset.layers['counts']In R: dataset$layers[["counts"]]
Type: sparsematrix, data type: float32, shape: 384925 × 41642
Raw counts
dataset.layers['normalized']In R: dataset$layers[["normalized"]]
Type: sparsematrix, data type: float32, shape: 384925 × 41642
Normalised expression values
dataset.obs['soma_joinid']In R: dataset$obs[["soma_joinid"]]
Type: vector, data type: int64, shape: 384925
If the dataset was retrieved from CELLxGENE census, this is a unique identifier for the cell.
dataset.obs['dataset_id']In R: dataset$obs[["dataset_id"]]
Type: vector, data type: category, shape: 384925
Identifier for the dataset from which the cell data is derived, useful for tracking and referencing purposes.
dataset.obs['assay']In R: dataset$obs[["assay"]]
Type: vector, data type: category, shape: 384925
Type of assay used to generate the cell data, indicating the methodology or technique employed.
dataset.obs['assay_ontology_term_id']In R: dataset$obs[["assay_ontology_term_id"]]
Type: vector, data type: category, shape: 384925
Experimental Factor Ontology (EFO:) term identifier for the assay, providing a standardized reference to the assay type.
dataset.obs['cell_type']In R: dataset$obs[["cell_type"]]
Type: vector, data type: category, shape: 384925
Classification of the cell type based on its characteristics and function within the tissue or organism.
dataset.obs['cell_type_ontology_term_id']In R: dataset$obs[["cell_type_ontology_term_id"]]
Type: vector, data type: category, shape: 384925
Cell Ontology (CL:) term identifier for the cell type, offering a standardized reference to the specific cell classification.
dataset.obs['development_stage']In R: dataset$obs[["development_stage"]]
Type: vector, data type: category, shape: 384925
Stage of development of the organism or tissue from which the cell is derived, indicating its maturity or developmental phase.
dataset.obs['development_stage_ontology_term_id']In R: dataset$obs[["development_stage_ontology_term_id"]]
Type: vector, data type: category, shape: 384925
Ontology term identifier for the developmental stage, providing a standardized reference to the organism’s developmental phase.
If the organism is human (organism_ontology_term_id == 'NCBITaxon:9606'), then the Human Developmental Stages (HsapDv:) ontology is used.
If the organism is mouse (organism_ontology_term_id == 'NCBITaxon:10090'), then the Mouse Developmental Stages (MmusDv:) ontology is used. Otherwise, the Uberon (UBERON:) ontology is used.
dataset.obs['disease']In R: dataset$obs[["disease"]]
Type: vector, data type: category, shape: 384925
Information on any disease or pathological condition associated with the cell or donor.
dataset.obs['disease_ontology_term_id']In R: dataset$obs[["disease_ontology_term_id"]]
Type: vector, data type: category, shape: 384925
Ontology term identifier for the disease, enabling standardized disease classification and referencing.
Must be a term from the Mondo Disease Ontology (MONDO:) ontology term, or PATO:0000461 from the Phenotype And Trait Ontology (PATO:).
dataset.obs['donor_id']In R: dataset$obs[["donor_id"]]
Type: vector, data type: category, shape: 384925
Identifier for the donor from whom the cell sample is obtained.
dataset.obs['is_primary_data']In R: dataset$obs[["is_primary_data"]]
Type: vector, data type: bool, shape: 384925
Indicates whether the data is primary (directly obtained from experiments) or has been computationally derived from other primary data.
dataset.obs['self_reported_ethnicity']In R: dataset$obs[["self_reported_ethnicity"]]
Type: vector, data type: category, shape: 384925
Ethnicity of the donor as self-reported, relevant for studies considering genetic diversity and population-specific traits.
dataset.obs['self_reported_ethnicity_ontology_term_id']In R: dataset$obs[["self_reported_ethnicity_ontology_term_id"]]
Type: vector, data type: category, shape: 384925
Ontology term identifier for the self-reported ethnicity, providing a standardized reference for ethnic classifications.
If the organism is human (organism_ontology_term_id == 'NCBITaxon:9606'), then the Human Ancestry Ontology (HANCESTRO:) is used.
dataset.obs['sex']In R: dataset$obs[["sex"]]
Type: vector, data type: category, shape: 384925
Biological sex of the donor or source organism, crucial for studies involving sex-specific traits or conditions.
dataset.obs['sex_ontology_term_id']In R: dataset$obs[["sex_ontology_term_id"]]
Type: vector, data type: category, shape: 384925
Ontology term identifier for the biological sex, ensuring standardized classification of sex. Only PATO:0000383, PATO:0000384 and PATO:0001340 are allowed.
dataset.obs['suspension_type']In R: dataset$obs[["suspension_type"]]
Type: vector, data type: category, shape: 384925
Type of suspension or medium in which the cells were stored or processed, important for understanding cell handling and conditions.
dataset.obs['tissue']In R: dataset$obs[["tissue"]]
Type: vector, data type: category, shape: 384925
Specific tissue from which the cells were derived, key for context and specificity in cell studies.
dataset.obs['tissue_ontology_term_id']In R: dataset$obs[["tissue_ontology_term_id"]]
Type: vector, data type: category, shape: 384925
Ontology term identifier for the tissue, providing a standardized reference for the tissue type.
For organoid or tissue samples, the Uber-anatomy ontology (UBERON:) is used. The term ids must be a child term of UBERON:0001062 (anatomical entity). For cell cultures, the Cell Ontology (CL:) is used. The term ids cannot be CL:0000255, CL:0000257 or CL:0000548.
dataset.obs['tissue_general']In R: dataset$obs[["tissue_general"]]
Type: vector, data type: category, shape: 384925
General category or classification of the tissue, useful for broader grouping and comparison of cell data.
dataset.obs['tissue_general_ontology_term_id']In R: dataset$obs[["tissue_general_ontology_term_id"]]
Type: vector, data type: category, shape: 384925
Ontology term identifier for the general tissue category, aiding in standardizing and grouping tissue types.
For organoid or tissue samples, the Uber-anatomy ontology (UBERON:) is used. The term ids must be a child term of UBERON:0001062 (anatomical entity). For cell cultures, the Cell Ontology (CL:) is used. The term ids cannot be CL:0000255, CL:0000257 or CL:0000548.
dataset.obs['batch']In R: dataset$obs[["batch"]]
Type: vector, data type: category, shape: 384925
A batch identifier. This label is very context-dependent and may be a combination of the tissue, assay, donor, etc.
dataset.obs['size_factors']In R: dataset$obs[["size_factors"]]
Type: vector, data type: float32, shape: 384925
The size factors created by the normalisation method, if any.
dataset.obsm['X_pca']In R: dataset$obsm[["X_pca"]]
Type: densematrix, data type: float32, shape: 384925 × 50
The resulting PCA embedding.
dataset.obsp['knn_connectivities']In R: dataset$obsp[["knn_connectivities"]]
Type: sparsematrix, data type: float32, shape: 384925 × 384925
K nearest neighbors connectivities matrix.
dataset.obsp['knn_distances']In R: dataset$obsp[["knn_distances"]]
Type: sparsematrix, data type: float64, shape: 384925 × 384925
K nearest neighbors distance matrix.
dataset.uns['dataset_description']In R: dataset$uns[["dataset_description"]]
Type: atomic, data type: str, shape: 1
Long description of the dataset.
dataset.uns['dataset_id']In R: dataset$uns[["dataset_id"]]
Type: atomic, data type: str, shape: 1
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
dataset.uns['dataset_name']In R: dataset$uns[["dataset_name"]]
Type: atomic, data type: str, shape: 1
A human-readable name for the dataset.
dataset.uns['dataset_organism']In R: dataset$uns[["dataset_organism"]]
Type: atomic, data type: str, shape: 1
The organism of the sample in the dataset.
dataset.uns['dataset_reference']In R: dataset$uns[["dataset_reference"]]
Type: atomic, data type: str, shape: 1
Bibtex reference of the paper in which the dataset was published.
dataset.uns['dataset_summary']In R: dataset$uns[["dataset_summary"]]
Type: atomic, data type: str, shape: 1
Short description of the dataset.
dataset.uns['dataset_url']In R: dataset$uns[["dataset_url"]]
Type: atomic, data type: str, shape: 1
Link to the original source of the dataset.
dataset.uns['knn']In R: dataset$uns[["knn"]]
Type: dict, data type: ``, shape: 3
Supplementary K nearest neighbors data.
dataset.uns['normalization_id']In R: dataset$uns[["normalization_id"]]
Type: atomic, data type: str, shape: 1
Which normalization was used
dataset.uns['pca_variance']In R: dataset$uns[["pca_variance"]]
Type: dict, data type: ``, shape: 2
The PCA variance objects.
dataset.var['soma_joinid']In R: dataset$var[["soma_joinid"]]
Type: vector, data type: int64, shape: 41642
If the dataset was retrieved from CELLxGENE census, this is a unique identifier for the feature.
dataset.var['feature_id']In R: dataset$var[["feature_id"]]
Type: vector, data type: object, shape: 41642
Unique identifier for the feature, usually a ENSEMBL gene id.
dataset.var['feature_name']In R: dataset$var[["feature_name"]]
Type: vector, data type: object, shape: 41642
A human-readable name for the feature, usually a gene symbol.
dataset.var['hvg']In R: dataset$var[["hvg"]]
Type: vector, data type: bool, shape: 41642
Whether or not the feature is considered to be a ‘highly variable gene’
dataset.var['hvg_score']In R: dataset$var[["hvg_score"]]
Type: vector, data type: float64, shape: 41642
A ranking of the features by hvg.
dataset.varm['pca_loadings']In R: dataset$varm[["pca_loadings"]]
Type: densematrix, data type: float32, shape: 41642 × 50
The PCA loadings matrix.