Info
openproblems_
Tasic et al. (2016)
1.91 GiB
02-02-2024
14249 × 34617
Adult mouse primary visual cortex
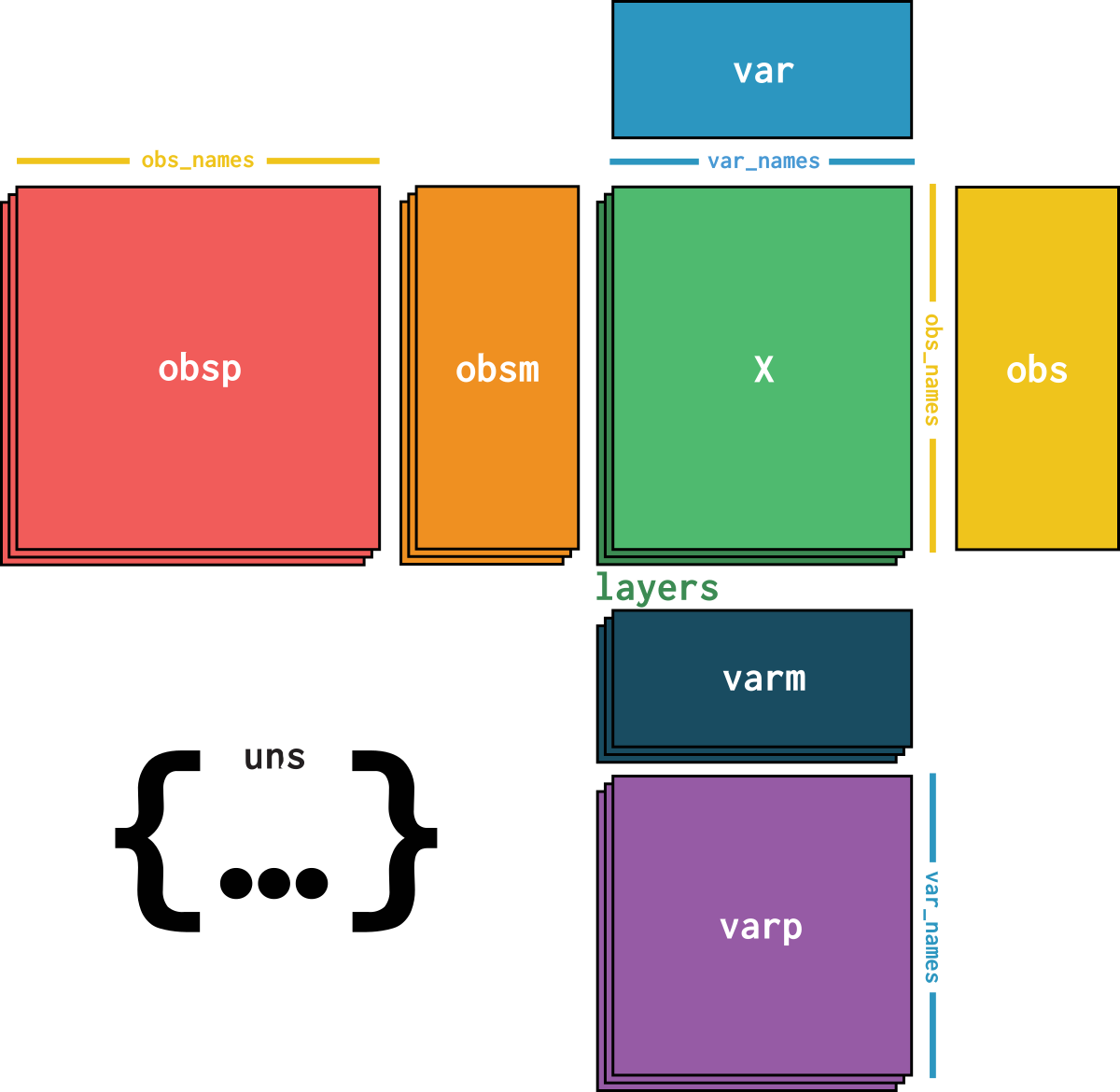
openproblems_
Tasic et al. (2016)
1.91 GiB
02-02-2024
14249 × 34617
CREATED
02-02-2024
DIMENSIONS
14249 × 34617
A murine brain atlas with adjacent cell types as assumed benchmark truth, inferred from deconvolution proportion correlations using matching 10x Visium slides (see Dimitrov et al., 2022).
dataset is an AnnData object with n_obs × n_vars = 14249 × 34617 with slots:
cell_type, size_factorsfeature_name, hvg, hvg_scoreknn_connectivities, knn_distancesX_pcapca_loadingscounts, normalizeddataset_description, dataset_id, dataset_name, dataset_organism, dataset_reference, dataset_summary, dataset_url, knn, normalization_id, pca_variance| Name | Description | Type | Data type | Size |
|---|---|---|---|---|
| obs | ||||
cell_
|
Classification of the cell type based on its characteristics and function within the tissue or organism. |
vector
|
category
|
14249 |
size_
|
The size factors created by the normalisation method, if any. |
vector
|
float32
|
14249 |
| var | ||||
feature_
|
A human-readable name for the feature, usually a gene symbol. |
vector
|
object
|
34617 |
hvg
|
Whether or not the feature is considered to be a ‘highly variable gene’ |
vector
|
bool
|
34617 |
hvg_
|
A ranking of the features by hvg. |
vector
|
float64
|
34617 |
| obsp | ||||
knn_
|
K nearest neighbors connectivities matrix. |
sparsematrix
|
float32
|
14249 × 14249 |
knn_
|
K nearest neighbors distance matrix. |
sparsematrix
|
float64
|
14249 × 14249 |
| obsm | ||||
X_
|
The resulting PCA embedding. |
densematrix
|
float32
|
14249 × 50 |
| varm | ||||
pca_
|
The PCA loadings matrix. |
densematrix
|
float64
|
34617 × 50 |
| layers | ||||
counts
|
Raw counts |
sparsematrix
|
float32
|
14249 × 34617 |
normalized
|
Normalised expression values |
sparsematrix
|
float32
|
14249 × 34617 |
| uns | ||||
dataset_
|
Long description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
|
atomic
|
str
|
1 |
dataset_
|
A human-readable name for the dataset. |
atomic
|
str
|
1 |
dataset_
|
The organism of the sample in the dataset. |
atomic
|
str
|
1 |
dataset_
|
Bibtex reference of the paper in which the dataset was published. |
atomic
|
str
|
1 |
dataset_
|
Short description of the dataset. |
atomic
|
str
|
1 |
dataset_
|
Link to the original source of the dataset. |
atomic
|
str
|
1 |
knn
|
Supplementary K nearest neighbors data. |
dict
|
3 | |
normalization_
|
Which normalization was used |
atomic
|
str
|
1 |
pca_
|
The PCA variance objects. |
dict
|
2 | |
dataset.layers['counts']In R: dataset$layers[["counts"]]
Type: sparsematrix, data type: float32, shape: 14249 × 34617
Raw counts
dataset.layers['normalized']In R: dataset$layers[["normalized"]]
Type: sparsematrix, data type: float32, shape: 14249 × 34617
Normalised expression values
dataset.obs['cell_type']In R: dataset$obs[["cell_type"]]
Type: vector, data type: category, shape: 14249
Classification of the cell type based on its characteristics and function within the tissue or organism.
dataset.obs['size_factors']In R: dataset$obs[["size_factors"]]
Type: vector, data type: float32, shape: 14249
The size factors created by the normalisation method, if any.
dataset.obsm['X_pca']In R: dataset$obsm[["X_pca"]]
Type: densematrix, data type: float32, shape: 14249 × 50
The resulting PCA embedding.
dataset.obsp['knn_connectivities']In R: dataset$obsp[["knn_connectivities"]]
Type: sparsematrix, data type: float32, shape: 14249 × 14249
K nearest neighbors connectivities matrix.
dataset.obsp['knn_distances']In R: dataset$obsp[["knn_distances"]]
Type: sparsematrix, data type: float64, shape: 14249 × 14249
K nearest neighbors distance matrix.
dataset.uns['dataset_description']In R: dataset$uns[["dataset_description"]]
Type: atomic, data type: str, shape: 1
Long description of the dataset.
dataset.uns['dataset_id']In R: dataset$uns[["dataset_id"]]
Type: atomic, data type: str, shape: 1
A unique identifier for the dataset. This is different from the obs.dataset_id field, which is the identifier for the dataset from which the cell data is derived.
dataset.uns['dataset_name']In R: dataset$uns[["dataset_name"]]
Type: atomic, data type: str, shape: 1
A human-readable name for the dataset.
dataset.uns['dataset_organism']In R: dataset$uns[["dataset_organism"]]
Type: atomic, data type: str, shape: 1
The organism of the sample in the dataset.
dataset.uns['dataset_reference']In R: dataset$uns[["dataset_reference"]]
Type: atomic, data type: str, shape: 1
Bibtex reference of the paper in which the dataset was published.
dataset.uns['dataset_summary']In R: dataset$uns[["dataset_summary"]]
Type: atomic, data type: str, shape: 1
Short description of the dataset.
dataset.uns['dataset_url']In R: dataset$uns[["dataset_url"]]
Type: atomic, data type: str, shape: 1
Link to the original source of the dataset.
dataset.uns['knn']In R: dataset$uns[["knn"]]
Type: dict, data type: ``, shape: 3
Supplementary K nearest neighbors data.
dataset.uns['normalization_id']In R: dataset$uns[["normalization_id"]]
Type: atomic, data type: str, shape: 1
Which normalization was used
dataset.uns['pca_variance']In R: dataset$uns[["pca_variance"]]
Type: dict, data type: ``, shape: 2
The PCA variance objects.
dataset.var['feature_name']In R: dataset$var[["feature_name"]]
Type: vector, data type: object, shape: 34617
A human-readable name for the feature, usually a gene symbol.
dataset.var['hvg']In R: dataset$var[["hvg"]]
Type: vector, data type: bool, shape: 34617
Whether or not the feature is considered to be a ‘highly variable gene’
dataset.var['hvg_score']In R: dataset$var[["hvg_score"]]
Type: vector, data type: float64, shape: 34617
A ranking of the features by hvg.
dataset.varm['pca_loadings']In R: dataset$varm[["pca_loadings"]]
Type: densematrix, data type: float64, shape: 34617 × 50
The PCA loadings matrix.